6-MOLECULAR BASIS OF INHERITANCE
CHAPTER NO.6 MOLECULAR BASIS OF
INHERITANCE
A51
INTRODUCTION
Hello students, in the previous chapter, you have
come to know, how the characters, pass on to their children. You are now
familiar with the word GENE.
Now in this chapter, “Molecular Basis of
Inheritance” we will come to know about the chemical or molecular nature of
GENES, which is the main genetic material in the world of GENETICS.
Genetic material is that substance which not only
controls the inheritance of traits from one generation to the next but is also
able to express its effect through the formation and functioning of the traits.
CHARACTERISTICS OF GENETIC MATERIAL:
A molecule that can act as a genetic material must
fulfil the following criteria:
1. It must be able to replicate,
2. Store information,
3. Express information, and
4. Allow variation by mutation.
Scientists worked hard to establish that hereditary
material is DNA in most
of organisms.
Meischer discovered NUCLEIN. He used this term for
DNA with associated proteins.
Mendel too had given the principles of inheritance
at same time and used the term ‘factor’ for genetic material.
By 1926, scientists had reached molecular level of
research to establish the identity of genetic material.
The famous scientists Frederick Griffith carried out
series of experiment with Streptococcus pneumoniae bacteria of S and R strain.
Mice injected with S strain developed pneumonia
whereas the mice injected with R strain did not.
Then he killed S strain bacteria with heat and
injected. Nothing happened, mice did not die.
Now he injected heat killed S strain and live R
strain bacteria. The mice
died.
This experiment led to thought believing DNA as
genetic material.
Many scientists worked to determine the biochemical
nature of ‘Transforming Principle’ in Griffith’s experiment.
Many scientists thought that genetic material was a
protein.
They extracted PROTEINS, DNA and RNA from heat
killed S cells.They discovered that Proteases and Rnase didn't affect
transformation but Dnase did inhibit transformation.
elt was concluded that DNA is the hereditary
material. Many scientists were
not convinced with this conclusion and many
continued their experim
PART:A VERY SHORT ANSWER TYPE QUESTIOS:
(i) Multiple Choice Questions:
1) Which of the following is false about
any molecule to be considered as genetic material?
(a) replicable
(b) inneritable
(c)non mutable
(d) all
2) Frederick Griffith carried out his
experiment with:
(a) Viruses
(b) Bacteria
(c) Bacteriophages
(d) none
3) How many strains of bacteria were used
in Griffith's experiment?
(a)2
(b)4
(c)3
(d)5
4) Meischer discovered:
(a) Nuclein
(b) Chromatin
(c) Histones
(d) None
5) On the culture plate, the rough colonies
produced by baceria of:
(a) M strain
(b) S strain
(c) R strain
(d) Z strain
(ii) True/ False:
1) The mice injected with live S strain bacteria
died from pneumonia.
2) Heat killed S strain bacteria when injected
alone, the mice died.
3) Genetic material snould be replicable.
(iii) Fill ups:
1) DNA is ............. acid.
2) .............. Strain is virulent Strain of
streptococcus bacteria
ANSWER KEY: PART-A
A) Multiple Choice Questions:
1) (c) non mutable It should be able to mutate so
that it may show
variations.
2) (b) Bacteria Griffith conducted a series of
experiments using Streptococcus
pneumoniae bacteria.
3) (a) Griffith used two strains of pneumococcus
(Streptococcus pneumoniae)
bacteria which infect mice — a type III-S (smooth)
which was virulent, and a type II-
R (rough) strain which was non-virulent.
4) (a) Nuclein
5) (c) R strain The R (rough) strain, lacks the coat
and produces colonies that
look rough and irregular.
B) True/ False:
1) True — The mice died due to virulent nature of S
strain bacteria.
2) False - The mice remained alive after injecting
heat killed S strain.
3) True - Genetic material should have the ability
to replicate so that it can make
its copy.
C) Fill Ups:
1) DNA is deoxyribose nucleic acid.
2) S Strain is considered as virulent strain as it
is responsible to cause disease.
PART: B_ SHORT ANSWER TYPE QUESTIONS:
1) Which biomolecules were considered to be possible
genetic material?
2) Enlist various characteristics of genetic
material.
3) Differentiate between S strain and R strains of
Streptococcus bacteria.
PART: C LONG ANSWER TYPE QUESTION:
1) Write the salient features of Griffith's
experiment.
A52
INTRODUCTION
Dear students, as in previous topic we have learnt
about Griffith's experiment
of Transformation, on mice.
TRANSFORMATION is defined as the change in genetic
material of an organism by the genes from outside as from remains of its dead
relatives.Griffth’s experiment however could not specify the Transforming
Chemical.Today we will study about experiments conducted by various scientists
to prove that DNA is the hereditary or genetic material.
A) AVERY & OTHERS EXPERIMENT:Oswald Avery, Colin
MacLeod and Maclyn McCarty (1933-44) purified biochemicals (protein, DNA, RNA,
etc.) from the heat killed S cells. They
discovered that DNA alone from S - bacteria caused R
- bacteria to become transformed.
They also discovered that transformation was not
affected by protein-digesting
enzymes (proteases), and RNA-digesting enzymes (RNases).
Hence, it was concluded that the transforming substance was not a
protein or RNA. Digestion with DNase inhibited transformation. This suggested
that the DNA was the cause of transformation. Thus, it was concluded that DNA
is the hereditary material.
Their findings were as under:
i) DNA alone from S- bacteria caused R- bacteria to
be transformed.
ii) They found that Proteases (protein digested
enzymes) and RNAase (RNA
digesting enzymes) did not affect transformation.
iii) Digestion with DNAase did inhibit
transformation.Thus they finally concluded that DNA is the hereditary material.
B) HERSHEY AND CHASE EXPERIMENT TO
PROVE DNA AS A GENETIC MATERIAL:
In 1952, Hershey and Chase conducted their blenders
experiment on T:phage which attacks the bacterium Escherichia coli.
1. The phage particles were prepared by using radio
isotopes of *S and *P .
2. These two radioactive phage preparations (one
with radioactive proteins and
another with radioactive DNA) were allowed to infect
the culture of E.coli.
3. The protein coats were separated from the
bacterial cell walls by shaking and
centrifugation. the heavier infected bacterial cells
during centrifugation pelleted to the bottom.
4. The supernatant had the lighter phage particles
and other components that
failed to infect bacteria.
5. It was observed that bacteriophages with
radioactive DNA gave rise to radioactive pellets *2P in DNA.
6. However in the phage particles with radioactive
protein (with °°S) the bacterial pellets have almost nil radioactivity,
indicating that proteins have failed to migrate into the bacterial cell.
So, it can be safely concluded that during infection
by bacteriophage T, it
was DNA which entered the bacteria. It was followed
by an eclipse period during which phage DNA replicates numerous times within
the bacterial cell.
Lysozyme (an enzyme) brings about the lysis of host
cells and releases the newly formed bacteriophages.The above experiment clearly
suggests that it is phage DNA and not protein which contains the genetic
information for the production of new bacteriophages.
PART:A VERY SHORT ANSWER TYPE QUESTIOS:
(i) Multiple Choice Questions:
1.) The initial work which formed the base
to establish DNA as genetic material was done by:
a.) Watson and Crick
b.) George Mendel
c.) Frederick Griffith
d.) Beadle and Tatum
2.) Which microorganism was involved in the
study of DNA as a genetic material by Griffith?
a.) S. pneumoniae
b.) Neurospora
c.) L. odoratus
d.) L. bacillus
3.) In Griffith's experiment, the
conversion of R-type to S-type of bacteria
when mixed with heat killed S-type is
called:
a.) Mutation
b.) Transduction
c.) Transfection
d.) Transformation
4.) The Blenders experiment to prove DNA as
genetic material was conducted by:
a.) Beadle and Tatum
b.) Frederick Griffith
c.) Hershey and Chase
d.) Hofmeister and Waldayer
5.) Which organism was allowed to infect
the culture of E.coli in Blender’s experiment?
a.) Bacteriophage
b.) Bacteria
c.) Algae
d.) Plasmodium vivax
(ii) Fill Ups:
1. | The chemicals used by Hershey and chase in
their experiment,were and.
2. The protein coats were separated from the
bacterial cell walls by and ;
(iii) True/False:
1. Hershey and Chase worked with Viruses that infect
bacteria in their experiment.
2. DNA alone from R- Bacteria, caused S-Bacteria to
be transformed.
ANSWER KEY: PART-A
(i) Multiple Choice Questions:
1. (c) Griffith performed transformation experiment
to prove that there is some genetic material which is responsible for carrying
characteristic functions
2. (a), Griffith used two strains of streptococcus
bacteria, S and R strain
3. (d), Transformation
4. (c), Hershey and Chase conducted blender’s
experiment which is also
known as Transduction.
5. (a) Bacteriophages are the viruses which infect
bacteria.
(ii) Fill Ups:
1. Radioactive Sulphur and Phosphorus
2. Shaking and centrifugation
(iii) True/False:
1. True
2. False: DNA alone from S- Bacteria caused
R-Bacteria to be transformed.
PART: B SHORT ANSWER TYPE QUESTIONS:
1. Name the scientists who proved Griffith's
experiment also DNA is the
genetic material.
2. What type of radioactive components were used in
Hershey and chase experiment?
3. Define transduction.
PART: C LONG ANSWER TYPE QUESTION:
1.) Discuss the contributions of Avery and
Co-workers in the study of DNA as
genetic material.
2.) How Hershey and Chase experimentally proved that
DNA is hereditary material?
A53
INTRODUCTION
WHAT IS DNA?
DNA- DEOXYRIBONUCLEIC ACID
DNA is a group of molecules that is responsible for
carrying and transmitting the hereditary materials or the genetic instructions
from parents to offspring.
It is an organic compound that has a unique
molecular structure.It is found in all prokaryotic cells and eukaryotic cells.
DNA STRUCTURE
The DNA molecule is composed of units called
nucleotides and each nucleotide is composed of three different components, such
as sugar, phosphate groups and nitrogen bases. When this unit lacks
DNA LOCATION
DNA is found in the cells of all the living
organisms except in some plant
viruses. In bacteriophages and viruses, there is a
central core of DNA which
is enclosed in a protein coat. In bacteria, and in
mitochondria and plastids of
eukaryotic cells, DNA is circular and lies naked in
the cytoplasm.
CHEMICAL COMPOSITION OF DNA:
Chemical analyses have shown that DNA is composed of
three different types of molecules.
1. Phosphoric acid (H3PO,4) has three reactive (-OH)
groups of which two are involved in forming the sugar phosphate backbone of
DNA.
2. Pentose sugar’ DNA contains 2’-deoxy-D-ribose (or
simply deoxyribose) which is the reason for the name deoxyribose nucleic acid.
3. Organic bases: The organic bases
are heterocyclic compounds containing nitrogen in
their rings; hence they are also called nitrogenous bases. DNA ordinarily
contains four different bases called Adenine (A),Guanine (G), Thymine (T) and
Cytosine (C).
CHARGAFF’s RULE According to this rule, the DNA
molecule should have an equal ratio of Pyrimidines (T and C) and Purines (A and
G). There is always equality in quantity between the bases A and T and between
the bases G and C. (Ais adenine, T is thymine, G is guanine, and C is
cytosine.)
A+tG=T+C
A=T and G=C
The number of purines and pyrimidines in the DNA
exist in the ratio 1:1.
It provides the basis of base pairing.
Withthe help of thisrule, one can determine the
presence of a base in the DNA and also determine the strand length.
Base ratio A+T/G+C is specific for a species but
remains constant.
IMPORTANCE OF CHARGAFF RULE:-Chargaff's
rules are important because they point to a kind of “grammar of biology”. a set
of hidden rules that gover the structure of DNA. This grammar ought to reveal
itself as patterns in DNA that are invariant across all species.
LET US KNOW WHAT WE HAVE LEARNT!
PART: A VERY SHORT ANSWER TYPE
QUESTIONS:
(a) Multiple choice questions:
(1) Which of the following options, are the
pyrimidine bases found in DNA?
(a) uracil and thymine
(b) thymine and cytosine
(c) adenine and thymine
(d) cytosine and uracil.
(2) A nucleoside differs from a nucleotide,
in lacking the:
(a) base
(b) sugar
(c) hydroxl group
(d) phosphate group
(3) The two strands in a DNA double is
joined by:
(a) covalet bond
(b) hydrogen bond
(c) ionic bond
(d) both a and b
(4)
DNA was first discovered by:
(a) J.D.watson
(b) Francis Crick
(c) Friedrich miescher
(d) H.G.khurana
(5) According to chargaff's rule, which one
is correct?
(a) A+T=G+C
(b) AtC=G+T
(c) A+G=T+C
(d) both a and c.
(b) Fill in the blanks:
(1) nitrogenous base is not present in DNA.
(2) New strand of DNA are found only in the
direction.
(3) A nucleotide consists of a a and a nitrogen
base.
(c) True /false:
(1) Adenine always pairs with cytosine.
(2) sugars and phosphates make up the backbone of
DNA.
(3) sugar in DNA is called ribose.
ANSWER KEY: PART -A
(a) MCQs:
(1) b--- thymine and cytosine are pyrimidine bases
found in DNA.
(2) d---- Phosphate group
(3) b----hydrogen bonds are responsible for base
pairing formation in DNA.
(4) c----friedrich miescher
(5) c ---A+G=T+C Acc. to chargaff rule any species
of any organism should have a 1:1 ratio of Purine and Pyrimidine bases.
(bi Fill Ups:
(1) Uracil -—-uracil nitrogenous base is present in
RNA.
(2) 5'-3'-—- new DNA is made by polymerase which
require template and
synthesize DNA in 5’3’ direction.
(3) Sugar, phosphate--—a nucleotide consist of
sugar, phosphate and nitrogen base.
ic) True/ false:
(1) False: (always pair with J acc. to Chargaff
Rule.
(2) True
(3) False; sugar in DNA is called deoxyribose.
PART: B SHORT ANSWER TYPE QUESTIONS:
(1) Which bases are purines and pyrimidines?
(2) write the chemical composition of DNA.
(3) Why is base pairing rule important?
PART: C LONG ANSWER TYPE QUESTIONS:
(1) If a double stranded DNA has 20% thymine
calculate the percentage of
guanine in the DNA.
(2) Explain the structure of D.N.A.
A54
INTRODUCTION
Students, in our previous topic we studied about
that DNA is universal genetic
material in all life forms. In our today’s topic we
will discuss its structure as
explained by Watson and crick. They proposed 3-Dimensional
model of DNA.
It was only in 1953 that James Watson and Francis
Crick, based on X-ray diffraction data produced by Maurice Wilkins and Rosalind
Franklin, proposed a very simple but famous double helix model for the
structure of DNA.
The three-dimensional structure of DNA, consists of
two long helical strands that
are coiled around a common axis to form a double
helix.
Each DNA molecule is comprised of two biopolymer
strands coiling around each
other.
Each strand has a 5’end (with a phosphate group) and
a 3'end (with a hydroxyl group).
The strands are antiparallel, meaning that one
strand runs in a 5'to 3'direction,
while the other strand runs in a 3'to 5'direction.
The diameter of the double helix is 2nm and the
double-helical structure repeats
at an interval of 3.4nm which corresponds to ten
base pairs.
The two strands are held together by hydrogen bonds
and are complementary
to each other.
The two DNA strands are called polynucleotides, as
they are made of simpler monomer units called nucleotides. Basically, the DNA
is composed of
deoxyribonucleotides.
The deoxyribonucleotides are linked together by 3-
5‘phosphodiester bonds.
The nitrogenous bases that compose the
deoxyribonucleotides include adenine, cytosine,
thymine, and guanine.
The structure of DNA -DNA is a double helix
structure because it looks like a twisted ladder.
The sides of the ladder are made of alternating
sugar (deoxyribose) and
phosphate molecules while the steps of the ladder
are made up of a pair of
nitrogen bases.
Asa result of the double-helical nature of DNA, the
molecule has two asymmetric grooves. One groove is smaller than the other.
This asymmetry is a result of the geometrical
configuration of the bonds between the phosphate,sugar, and base groups that forces
the base groups
to attach at 120-degree angles instead of 180
degrees.
The larger groove is called the major groove, occurs
when the backbones are far apart; while the smaller one is called the minor
groove, and occurs when they are close together.
Since the major and minor grooves expose the edges
of the bases, the grooves can be used to tell the base sequence of a specific
DNA molecule.
The possibility for such recognition is critical
since proteins must be able to
recognize specific DNA sequences on which to bind in
order for the proper functions of the body and cell to be carried out.
The Watson and Crick model shows that DNA is a
double helix with sugar-phosphate backbone on the outside and paired bases on
the inside.
PART: A VERY SHORT ANSWER TYPE
QUESTIOS:
(i) Multiple Choice Questions:
1. The two strands in DNA are coiled to
each other:
(a) Parallel
(b) anti parallel
(c) both a) and b)
(d) none of these
2. The nucleic acids are polymer of:
(a) amino acids
(b) amides
(c) nucleotides
(d) phosphates
3. Which of the following combination of
base pair is absent in DNA?
(a) A-T
(b) A-U
(c) C-G
(d) T-A
4. In a polynucleotide chain, nucleotide
are held through
(a) peptide linkage
(b) glycosidic linkage
(c) phosphodiester linkage
(d) hydrogen bond
5. A nucleotide is formed by the
combination of
(a) phosphate and sugar
(b) sugar phosphate
(c) base-sugar- phosphate
(d) none of these
(ii) True/ False:
1. 8base pairs are present in one full tum of
DNA-helix.
2. Base pairing pattern of DNA is A-U and G-C
3. Two strands of DNA molecule are held together by
phosphodiester bond.
(iii) Fill ups:
1. The DNA molecule takes a complete turn after
..........0.00..... base
pairs.
2. Anucleoside combines with......................to
form ucleotide.
ANSWER KEY: PART-A
A) Multiple Choice Questions:
1. (b) anti parallel It is because two chains are
parallel but their 5’->3
directions are opposite
2. (c) nucleotides A nucleotide consists of a base,
a sugar molecule and a
phosphate molecule.
3. (b) A-U Because Uracil is present in RNA
4. (c) phosphodiester linkage
5. (c) base-sugar-phosphate
B) True/ False:
1) False - 10 base pairs are present
2) False - the base pair pattern is A-T and G-C
3) False - The hydrogen bonds between complementary nucleotides
keep
two. strands of DNA helix together.
C) Fill Ups:
1) 10 base pairs
2) Phosphate group
PART: B SHORT ANSWER TYPE QUESTIONS:
1. Distinguish between nucleosides and nucleotides.
2. The two strands of DNA molecule are
anti-parallel. What do you understand by
anti-parallel?
3. What are the four pairs of DNA bases that form in
the double helix?
PART: C LONG ANSWER TYPE QUESTION:
1. Describe Watson and Crick model of DNA with the
help of well labelled
diagram.
A55
INTRODUCTION
DNA, the genetic information carrier molecule of the
cell is a long polymer
of nucleotides and can adopt different types of
structural conformations
the various types of conformations that the DNA can
adopt depend upon different factors such as:-
Hydration level
Salt concentration
DNA sequence
Quantity and direction of super-coiling
Presence of chemically modified bases
Presence of polyamines in solution
DIFFERENT
FORMS OF DNA:
1) A-DDNA
2)B-DNA
3)C-DNA
4)D-DNA
5)Z-DNA
A-DNA:A DNA is a rare type of structural
conformation, which can adopt under
dehydrating condition
MAIN FEATURES OF A-DNA -
A-DNA has shorter and more compact structure
organisation
A-DNA was discovered by Rosalind Franklin
A-DNA is formed from B DNA under dehydrating
condition
A-DNA is much wider and flatter than B-DNA
A-DNA is a right handed helix
The helix diameter of A-DNA is 26A
A-DNA containing 11.6 base per turn
A-DNA has narrow and deep major groves
The minor groves of A-DNA are wide and shallow
B-DNA
B-DNA is the most common type of DNA present in the
cell it is also called
biological DNA .B-DNA was discovered by James Watson
and Francis Crick.
MAIN FEATURES OF B-DNA:-
Majority of the DNA in a cell is B DNA.
B-DNA is a right-handed helix.
The helical diameter of B-DNA is 20 A each tum in
the B-DNA consists of 10 base pairs.
The B-DNA has a solid Central core.
The major grove of B-DNA is wide and deep the minor
grove of B-DNA is narrow and deep.
C-DNA
C DNA has 9.33 base pairs which are negatively
tilted from the axis of the
helix it's helical diameter is 19 A.
D-DNA
D-DNA has 8 base pairs which are negatively tilted
from the axis of the helix.
Z-DNA
Z DNA is a left-handed double helical confirmation
of DNA in which the
double helix winds to the left in zigzag pattern, it
was discovered by Andres
Wang and Alexander Rich.
MAIN FEATURES OF Z-DNA-
The helical diameter of z DNA is 18 A, with 12 base
pair.
LET US KNOW WHAT WE HAVE LEARNT: -
PART A- VERY SHORT ANSWER TYPE
QUESTIONS: -
(1) MULTIPLE CHOICE QUESTIONS:
Q1: The
helical diameter of B-DNA is:
a) 30 A°
b) 21 A°
c)32 A°
d)20 A°
Q2: Major groove of B DNA is:
a) Wide and deep
b) Shallow and deep
c) Narrow and wide
d) Narrow and deep
Q3: Name the three DNA that occur under
dehydration conditions:
a) A-DNA
b) B-DNA
c) C-DNA
d) Z-DNA
Q4: Left-handed DNA is:
a) A-DNA
b) B-DNA
c) Z-DNA
d) C-DNA
Q5: The helical diameter of Z- DNA is:
a) 12 A°
b) 30 A°
c) 28 A°
d) 18 A°
(i) FILL UPS
a) Z-DNA was discovered by.........
b) A-DNA contains........ base pair per turn
c) B-DNA is called........ DNA
(ii) TRUE/ FALSE
a) Helical diameter of a DNA is 26 A’.
b) Each turn in B-DNA consists of 12 base pair.
c) B-DNA was discovered by James Watson and Francis
Crick.
ANSWER KEY: PART-A
(i) MULTIPLE CHOICE QUESTIONS: -
1) (d) 20A
2) (a) wide and deep
3) (a) A-DNA- as it can adopt under dehydrating
conditions.
4) (d) Z-DNA- as its double helix winds in a zig-zag
pattern
5) (d) 18A
(ii) FILL UPS: -
1) Andres Wang and Alexander Rich
2) 11.6 base pairs
3) Biological DNA- as it is the most common type of
DNA found in the cell
(ii) TRUE FALSE:-
1) TRUE
2) FALSE
3) TRUE
PART: B SHORT ANSWER TYPE QUESTIONS:-
1) Define DNA a molecule
2) Name different forms of DNA
3) Give main features of B-DNA
PART: C LONG ANSWER TYPE QUESTIONS:-
1) Give difference between A-DNA, B- DNA and Z-DNA
A56
INRODUCTION
The DNA or deoxyribonucleic acid is not only the
largest negatively charged
macromolecule but also represents genetic material
of organism and molecular basics of heredity. The most important property of
DNA is super coiling.In Prokaryotic cells, DNA is circular and embedded in the
cytoplasm and is often called nucleoid. It is not bounded by nuclear membrane
and is without histone proteins. It is termed as naked DNA.Many Prokaryotes
also possess extra chromosomal small circular DNA
segments called Plasmids.
In Eukaryotic cells, DNA is linear and mainly
confined to the nucleus as the
component of chromosomes. It is termed as nuclear
DNA. It is associated with histone proteins to form chromatin fibres.
A small quantity of DNA is also present in the
mitochondria and plastids which is termed as extra nuclear or organellar DNA.
It is circular like prokaryotic DNA.
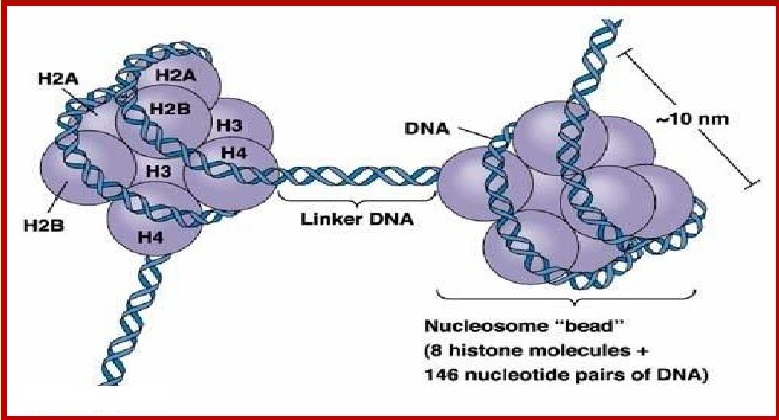
PACKAGING OF DNA
The length of the DNA is around 3 meters that need
to be accommodates within the nucleus which is only a few micrometres in
diameter.
In order to fit- in, the DNA molecules into the
nucleus, DNA,needs to be packed into an extremely compressed and compact
structure called chromatin.
DNA is condensed into a complex structure with
histone and non-histone proteins. Histones are rich in lysine and arginine and
are positively charged that provides structural support to a chromosome.
Histone proteins give a more compact shape to
chromosomes. Positive charge of histones allows them to associate with
negatively charged DNA.
There are five types of histones named H1, H2A, H2B,
H3 and H4.
Histones form a unit of eight molecules called
histone octamer at a centre
of a nucleosome core particle. The negatively charged
DNA is wrapped
around the positively charged histone octamer to
form a structure called
Nucleosome.
Linker DNA is double-stranded DNA 38-53 bp long
inbetween two nucleosome cores that, in association with histone H1, holds the
cores together. Linker DNA is seen as the string in the "beads and string
model"
Nucleosome contribute thread like coloured bodies in
the nucleus called chromatin. The nucleosomes in chromatin are seen like beads
on string.
Chromatin exists in two forms Euchromatin and Heterochromatin.
Euchromatin is lightly stained and loosely packed
chromatin whereas
Heterochromatin is darkly stained and more densely
packed chromatin.
LET US KNOW WHAT WE HAVE LEARNT!
PART-A VERY SHORT ANSWER TYPE
QUESTIONS:
MCQ’s
1. DNA is not present in:
(a) Nucleus
(b) Chloroplast
(c) Ribosomes
(d) Mitochondria
2. DNA fragments are:
(a) Negatively charged
(b) Either positively or negatively charged
(c) Positively charged
(d) Neutral
3. A nucleosome is best described as:
(a) One fully packaged DNA molecule
(b) One strand of DNA double helix
(c) Nucleosomes coiled around each other
(d) ADNA strand wrapped around histone proteins
4. What are the set of positively charged
basic proteins called as?
(a) Histidine
(b) DNA
(c) RNA
(d) Histones
5. When the negatively charged DNA combines
with positively charged histone octamer, which of the following is formed?
(a) Nucleus
(b) Nucleoid
(c) Nucleosome
(d) none
FILL UP’s >
1. Histones are rich in and amino acids.
2. The positively charged basic proteins are called
.
TRUE/FALSE >
1. Histones are negatively charged.
2. The length of DNA is near about 3m.
ANSWER KEY: PART-A
MCQ’s >
1. (c) Ribosomes are present in cytoplasm as well as
matrix of Mitochondria and Chloroplast. They do not contain any DNA.
2. (a) DNA is Negatively charged because of the
presence of one negatively charged oxygen in phosphate group.
3. (c) ADNA strand wrapped around histone proteins.
4. (d) Histones are the basic proteins because they
are rich in basic amino acids i.e. Arginine and lysine.
5. (c) Nucleosome
FILL UP’s
1. Lysine, Arginine
2. Histones
TRUE/FALSE
1. False Histones are the basic proteins because
they are rich in basic amino acids i.e. Arginine and lysine. These are
positively charged.
2. True
PART-B SHORT ANSWER TYPE QUESTIONS:
1. Whatis linker DNA and what is its function?
2. What type of appearance is given by nucleosome
chain under electron
microscope?
PART-C LONG ANSWER TYPE QUESTIONS:
1. What are the reasons of stabilization of
Histone-DNA interaction?
A57
INTRODUCTION
In molecular biology, DNA replication is the
biological process of producing two identical replicas of DNA from one original
DNA molecule.DNA replication occurs in all living organisms acting as the most
essential part for biological inheritance.
DNA Replication is Semi Conservative:
DNA replication is a semi conservation process
because when a new double
stranded DNA molecule is formed, one strand will be
form original template molecule and one strand will be newly synthesised. That
is half strand of DNA is conserved.
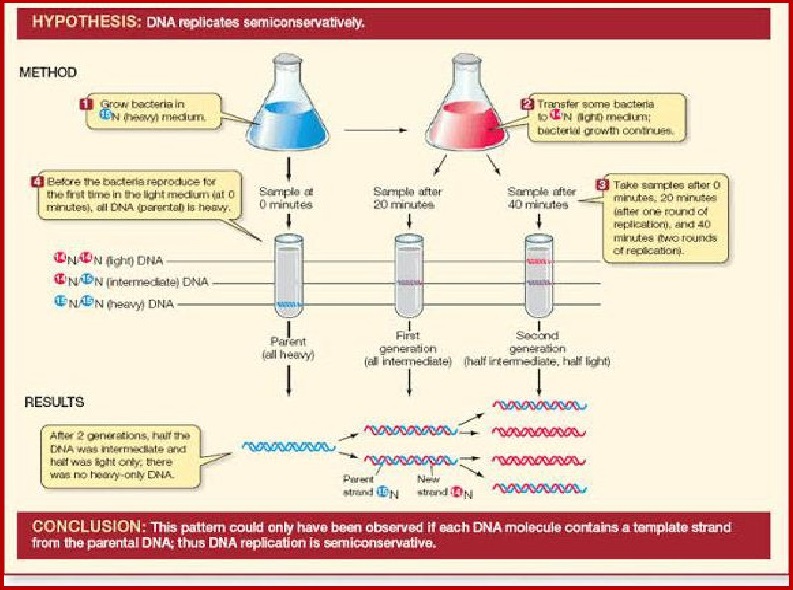
Meselson and Stahl Experiment to Prove that DNA
Replication is Semiconservative in Nature:Matthew Meselson and Franklin Stahl
performed the following experiment in 1958:
AIM : To prove the
mode of DNA replication is semi conservative
EXPERIMENTAL MATERIAL:
E. coli, lsotopes of nitrogen N-15, N-14
PRINCIPLE : Density gradient
centrifugation
1. E. Coli grown in ‘*N medium:They
grew E. coli ( Escherichia coli) in a medium containing '°NH.C!l ('°N is the
heavy isotope of nitrogen)as the only nitrogen source for many generations. The
result was that 'N was incorporated into newly
synthesised DNA.
This heavy DNA molecule could be distinguished from
the normal DNA by centrifugation in a cesium chloride (CsCl) medium or
densitygradient.
Being '°N not a radioiosotopic isotope, it can be
separated from '4N only based on densities.
2. Transformation of N cell to N
medium.
Then they transferred the cells into a medium with
normal NH.Cl and took
samples at various definite time intervals as the
cell multiplied and extracted the
DNA that remained as double-stranded helices. The
various samples were
separated independently on CsCl gradients to measure
the densitiesof DNA.
3. Replication.
After that the DNA was extracted from the culture,
one generation after the
transfer from '°N to '4N medium , that is after 20
minutes the cells of E.coli
divides and had a hybrid or intermediate density.
DNA extracted from the
culture after another generation that is after 40
minutes, 2"? generation
composed of equal amount this hybrid DNA and of
light DNA.
CONCLUSION
Meselson and Stah! concluded that DNA replication is
not conservative but
Semi-Conservative.
Each strand in DNA molecule serves as a template for
synthesis of new
complementary strand.
Each strand is composed of one parent strand and one
newly synthesized
strand.
LET US KNOW WHAT WE HAVE LEARNT!
PART:A VERY SHORT ANSWER TYPE
QUESTIONS;
(i) MULTIPLE CHOICE QUESTIONS:
1) Semiconservative mode of replication of
DNA was proved by:
(a)Hershey and Chase
(b) Griffith
(c) Watson and Crick
(d) Meselson and Stahl
2) Semiconservative replication of DNA was
first demonstrated in:
(a)Drosophila melanogaster
(b) Escherichia coli
(c) Streptococcus pneumonia
(d) Salmonella typhi
3) Which process was used by Meselson and
Stahl for their experiment:
(a)Centrifugation
(b) Chromatography
(c) Density gradient centrifugation
(d) Buoyant density centrifugation
4) Which isotope out of the following was
used:
(a) N
(b) O'8
(c)C"4
(d ) all of these
5) Which medium is used for centrifugation
in the experiment:
(a) cesium chloride
(b) sodium chloride
(c) ammonium chloride
(d) potassium chloride
(ii) FILL UPS :
1) The Meselson and Stahl experiment is based on the
principle
2) This experiment proves that DNA replication is
3) The semiconservative replication of DNA was
proved in year
(iii) TRUE/FALSE:
1) E. coli divides in 30 minutes
2) Ammonium chloride (NHsCL) is the only source of
nitrogen in the experiment
ANSWER KEY: PART-A
A) MULTIPLE CHOICE QUESTIONS :
1) (d) Meselson and stahl they proved the
semiconservative mode of replication .
2) (b) Escherichia coli as in E.coli replication
occurs with high accuracy
3) (c) Density gradient centrifugation — This type
of centrifugation separates
different particles based on their different
densities.
4) (a) N'* — They use heavy isotope.
5) (a) Cesium chloride — It is a density gradient.
B) FILL UPS:
1) Density gradient centrifugation
2) Semi conservative
3) 1958
C) TRUE / FALSE
1) False — E-coli divides in 20 minutes
2) True
PART-B SHORT ANSWER TYPE QUESTIONS:
1) What are the requirements of Meselson and Stahl
experiment?
2) Why did Meselson and Stahl use nitrogen?
3) What is semi conservative DNA replication? By
whom was is experimentally proved?
PART-C LONG ANSWER TYPE QUESTION:
1) Describe Meselson and Stahl’s experiment to show
that DNA replication is semi conservative.
A58
INTRODUCTION
Students, in previous topic, we have studied about
the properties of genetic
material. Also in the Meselson and Stahl’s
experiment we have come to know
that replication of DNA is emiconservative. In our
today’s topic we will discuss
DNA replication process in detail.
DNA replication is a process by which DNA make copy
of itself during cell-
division. It is a semi conservative process in which
one strand of the DNA is
parental and other one is newly synthesized. It is
an important process that takes place in the dividing cells. The cell undergoes
DNA replication during the S-phase of the cell cycle.
GENERAL FEATURES OF DNA REPLICATION:
1. DNA replication is semi conservative.
2. It is bidirectional process.
3. It proceeds from specific point called origin of
replication.
4. It proceeds from 5’ to 3’ direction.
5. It is multi enzymatic process.
6. It occurs with high degree of fidelity.
When DNA replicates, various types of enzymes are
involved. These enzymes
have the ability to quicken the reaction by building
or break down the items that
they act upon. Below listed are the enzymes involved
in DNA replication:
|. Helicase: It opens the DNA helix by breaking
hydrogen bonds between the
nitrogenous bases.
Il. Topoisomerase: It helps to relieve the stress on
DNA when unwinding by
causing breaks and then resealing the DNA.
Ill. DNA polymerase I: It undergoes exonuclease
activity by removing RNA primer
and replaces it with newly synthesized DNA.
IV. DNA polymerase Il: It repairs DNA.
V. DNA polymerase Ill: It is the main enzyme that
adds nucleotides in the 5-3’
direction.
VI. RNA primase: It synthesize RNA primers needed to
start the replication.
Vil. Ligase: It seals the gaps between the Okazaki
Fragments to create one
continuous DNA strand.
VII. Single strand binding proteins (SSB): It binds
to single stranded DNA to avoid
DNA rewinding back.
DNA replication involves following
steps: -
1. ACTIVATION OF DEOXYRIBONUCLEOTIDES -
The four nucleotides of DNA i.e., AMP, GMP, CMP & TMP are floating in
nucleus. They are
activated by ATP to form deoxyribonucleosides triphosphates called,
ATP, GTP, CTP & TTP.
2. RECOGNITION OF INITIATING POINTS
From a particular point unwinding of DNA molecule start with the help of
specific initiator proteins.
3. UNWINDING OF DNA MOLECULE —
DNA double helix unwind into single strands by the breakdown of weak hydrogen
bonds. Unwinding of Helix is helped by enzyme helicase. The separation of two
single strands of DNA
creates Y-shaped replication fork.
4. FORMATION OF RNA PRIMER -
The DNA directed RNA polymerase form the RNA primer.
5. FORMATION OF NEW DNA CHAINS The enzymes DNA
polymerase can polymerize the nucleotides only in 5’ 3’ direction. DNA
polymerase is responsible for the template direct condensation of
deoxyribonucleotide
triphosphates.
One of the strands is oriented in the 3’ > 5
direction is called leading strands. The other strand oriented in the 5’ — 3'
direction called lagging strands. As the results of their different
orientation, the two strands are replicated differently.
A short piece of RNA called a PRIMER comes along and
binds to the end of the leading strand. The primer act as the starting point
for the DNA synthesis.
DNA polymerase binds to the leading strand and then
walks along it,adding new complimentary nucleotide basis (A, C, G & T) to
the strands of DNA in the 5’—.3’ direction.
This sort of replication is called continuous.
lagging strand:Small pieces of DNA called Okazaki
fragments are than added to the lagging strand in 5’ — 3’ direction.
This type of replication is called Discontinues as
the Okazaki fragments,
formed in parts, will need to be joined up.
6. REMOVAL OF RNA PRIMER —-
Once the small pieces of Okazaki fragments have been formed RNA primer is
removed from 5’ end.
7, PROOF READING-
Proof Reading is done to make sure there is no mistake in the new DNA
sequence.Finally, an enzyme called DNA ligase seals up the sequence of DNA in
the
two continuous and discontinuous double strands.
LET US KNOW WHAT WE HAVE LEARNT!
PART:A_VERY SHORT ANSWER TYPE QUESTIOS:
(i) Multiple Choice Questions:
1. What is the nature of DNA replications?
(a) Conservative
(b) Non-Conservative
(c) Semi-Conservative
(d) None
2. The Unwinding of Helix is helped by
enzymes:
(a) Helicase
(b)Topoisomerases
(c) Polymerase
(d) Ligase
3. Which of the following is true about DNA
polymerase?
(a) It can synthesize DNA in the 5’ — 3’ direction
(b) ) It can synthesize DNA in the 3’ — 5’ direction
(c) It can synthesize mRNA in the 3’ — 5’ direction
(d) It can synthesize MRNA in the 5’ — 3’ direction
4. Okazaki fragments are:
(a) Large
(b) Small
(c) Both type
(d) None
5. Which of the following reaction is
required for proof reading during
DNA replication by DNA polymerase?
(a) 5° — 3' Exonuclease activity (b) 3’ — 5’
Exonuclease activity
(c) 3’ -+ 5 Endonuclease activity (d) 5° — 3’
Endonuclease activity
(ii) True/ False:
1. Unwinding of Helix is helped by Enzyme helicase.
2. The four nucleotides of DNA are ATP, GTP, CTP
& TTP.
(iii) Fill ups:
1. RNAprimer is removed at ..............0......
end.
2. DNA directed ...................... polymerase
forms the RNA primer.
ANSWER KEY: PART-A
A) Multiple Choice Questions:
1. (c) Semi-Conservative — Each DNA strand serve as
a template for the synthesis of a new strand producing two new DNA molecule,
each with one new strand and one old strand.
2. (A) Helicase - A DNA helicase is an enzyme that
unwind the DNA double helix
by breaking the hydrogen bounds between the
complimentary basis.
3. (A) It can synthesize DNA in the 5’ — 3’
direction — DNA pole can synthesize only anew DNA strandin 5’ — 3 direction
only.
4. (B) Small- Okazaki fragments are small sequence
of DNA nucleotides (Approximately 150 to 200 base pair in eukaryotes)
5. (B) 3’ — 5 Exonuclease activity- This removes the
mis- paired nucleotides
and polymerase begins again activities known as
proof reading.
B) True/ False:
1) True
2) False - The four nucleotide of DNA are AMP, GMP,
CMP & TMP.
C) Fill Ups:
1) 5
2) RNA
PART: B SHORT ANSWER TYPE QUESTIONS:
1. What are Okazaki fragments?
2. What do you mean by leading strands?
PART: C LONG ANSWER TYPE QUESTION:
1. Explain the process of DNA replication?
A59
INTRODUCTION
Students, in our previous topic we have learnt that
DNA, or deoxyribonucleic
acid, is the hereditary material in humans and
almost all other organisms. Here in
today’s topic, we will study about another important
nucleic acid which is responsible for converting the genetic information of DNA
into Proteins.
RNA or Ribose Nucleic Acid is the other nucleic acid
present in all biological
cells. It is principally involved in the synthesis
of protein, carrying the messenger instructions from DNA which itself contains
the genetic instruction required for the development and maintenance of life.
STRUCTURE OF RNA-
RNA is unbranched single- stranded polymer of
ribonucleotides.
Each nucleotide unit is composed of three molecules:
a phosphate group, a 5-carbon ribose sugar, and a nitrogen containing base.
The bases in RNA are Adenine(A), Guanine(G),
Uracil(U) and Cytosine(C).
The various components are linked up as in DNA i.e.
Adenine bonds with Uracil and Cytosine bonds with Guanine.
TYPES OF RNA:-
There are three types of RNA in every cell.
The three types of RNA are transcribed from
different regions of DNA template.
The types of RNA are classified into three types:
1. mRNA or messenger RNA
2. tRNA or transfer RNA
3. rRNA or ribosomal
RNA
1. MESSENGER RNA or mRNA:
The DNA that controls protein synthesis is located
in the chromosomes within the nucleus.
The ribosome, on which the protein synthesis
actually occurs are placed in
the cytoplasm.
Therefore, some sort of agency must exist to carry
instructions from the DNA to ribosomes. The agency does exist in the form mRNA.
The mRNA carries the message (information) from DNA
about the sequence of particular amino acid to be joined to form a polypeptide,
hence its name, messenger RNA.
Itis also called information RNA or template RNA.
2. TRANSFER RNA or t RNA:
The tRNA has many varieties.
Each variety carries a specific amino acid from the
amino acid pool to the mRNA on the ribosomes to form a polypeptide, hence, its
name.
The tRNA form about 15% of the total RNA of a cell.
Its molecule is the smallest of all the RNA types.
A tRNA molecule, as proposed by R.W, Holley in 1965,
has the form of a clover leaf that results from self-folding and base pairing
creating paired stems and unpaired loops.
Ithas four regions:
1. Amino acid carrier end
2. Anticodon end
3. DArm or Enzyme Site
4. TyC Am or Ribosome Site
3. RIBOSOMAL RNA or r RNA: -
The rRNA molecule is greatly coiled.
In combination with protein, it forms the small and
large sub-units of the ribosomes, hence its name.
It forms about 80% of the total RNA of a cell.
A eukaryotic ribosome is 80S.
Its large 60S subunit consists of 28S, 58S and 5S
rRNAs and over 45 different basic proteins.
Its small 40S subunit comprises 18S RNA and about 33
different basic proteins.
A prokaryotic ribosome is 70S.
Its large 50S subunit consists of 23S and 5S rRNAs
and about 34 different basic proteins.
Its small 30S subunits comprise 16S rRNA and about
21 different basic proteins.
The 3' end of 18S rRNA (16S rRNA in prokaryotes) has
a binding site for the
mRNA Cap.
The 5S rRNA has a binding site for t RNA.
ADDITIONAL RNA TYPES: -
There are two more minor types of RNAs
1. Small nuclear RNA or snRNA
2. Small Cytoplasmic RNA or scRNA
SnNRNA Role- It helps in processing of rRNA and mRNA
in the nucleus.
ScRNA Role-lt helps in binding the ribosomes to
endoplasmic reticulum (ER) in the cytoplasm.
All the three kinds of RNA play an important role in
PROTEIN SYNTHESIS.
Messenger RNA (mRNA), carries the genetic
information copied from DNA.
Transfer RNA (tRNA) is the key to deciphering the
code words in MRNA.Each type of amino acids has its own type of tRNA, which
binds it and carries it to the growing end of a polypeptide chain if the next
code word on MRNA calls for it.
Ribosomal RNA (rRNA) associates with a set of
proteins to form
ribosomes.
RNA serves as genetic material in some viruses.
Example: Tobacco Mosaic Virus (TMV)
RNA carries out a broad range of functions, from
translating genetic
information into the molecular machines and
structures of the cell to
regulating the activity of genes during development,
cellular differentiation,
and changing environments.
PART: A VERY SHORT ANSWER TYPE
QUESTIONS:
(i) MULTIPLE CHOICE QUESTIONS:
1. How many types of main RNA present in
eukaryotes?
a) 1
b) 2
c) 3
d) 4
2. Which RNA carries information from
nucleus to cytoplasm?
a) rRNA
b) mRNA
c) tRNA
d) ScRNA
3. Who proposed the clover leaf shape of
tRNA?
a) T.H morgan
b) George John Mendel
c) R.W Holley
d) Watson & crick
4. RNA acts as genetic material in which
types of virus?
a) Potato blight
b) Salmonela typhi
c) Corona
d) Tobbaco mosaic virus (TMV)
5. What is the percentage of rRNA in total
RNA?
a) 15%
b) 4%
c) 80%
d) 70%
(ii) FILL Ups:
1. RNAserveras _________ Material in Some Virus
(TMV)
2. ThemRNA has______ SHAME Of Molecule.
(iii) TRUE / FALSE:
1. mRNAis also known as template RNA.
2. R.W. Holley in 1965 told that tRNA has clover
leaf form.
I. MCQs:
1. (c) There are three types of RNA: mRNA, tRNA,
rRNA
2. (b) Messenger RNA (mRNA) decodes the information
of DNA by Transcription process in nucleus.
3. (c) R.W. Holley
4. (d) Tobbaco Mosaic virus (TMV) contains single
stranded RNA as genetic material.
5. (c) rRNA constitutes approx. 80% of the total RNA
in a cell.
Il. FILL Ups:
1. Genetic
2. Linear
lll. TRUE /FALSE:
1. True
2. True
PART: B SHORT ANSWER TYPE QUESTIONS:
1Q. Write the different types of RNA?
2Q. Write the difference between MRNA & tRNA?
3Q. Write the role & importance of MRNA?
4Q. Describe TMV?
5Q. Write the structure & role of RNA?
PART: C LONG ANSWER TYPE QUESTIONS:
Q1. Describe the various types of RNA in details?
A60
INTRODUCTION
Dear students, as in previous topic we have learnt
about DNA replication.DNA replication is the process by which a double-stranded
DNA molecule is
copied to produce two identical DNA molecules.Today
we will study about transcription.The process of copying or transferring
genetic information from one strand of the DNA into MRNA is termed as
transcription.
TRANSCRIPTION
This process occurs in nucleus in the presence of an
enzyme, RNA polymerase.
In other words,The process of synthesis of mRNA from
DNA or flow of genetic information from DNA to mRNA is known as transcription.
Where, Why and When transcription takes place?
We know, DNA is present within the nucleus and
protein synthesis occurs in cytoplasm.
Total raw material for synthesis of protein is
present in cytoplasm
The whole biological information about the type of
protein, which is going to form, is present in DNA.
Being larger in size DNA can not come outside
nucleus through nuclear pores.
That is why DNA transfers its information to RNA
(messenger RNA or mRNA).
This process occurs in G1 and G2 phase of interphase
of cell cycle.Process of transcription:
General Information:
DNA is double stranded.
Biological information is present in one strand and
it is Known as sense or
coding strand. This strand lies in the sequence of
5’ — 3’.
Its complementary strand is known as antisense
strand and lies in sequence of 3’ — 5’. It acts as template for the formation
of RNA.
RNA formed on template strand will be in the
sequence of 5’ — 3’.
In the nucleotide of RNA, nitrogenous base URACIL is
present instead of nitrogenous base THYMINE as compared to nucleotide of DNA.
RNA polymerase enzymes are required for this
process.
RNA polymerase is only of one type in prokaryotes.
It has two extra factors i.e. sigma (o) factor
(helps in initiation of
transcription) and rho (p) factor (helps in
completion or termination of transcription).
RNA polymerase in eukaryotes is of three types
RNA polymerase | (forms ribosomal or rRNA)
RNA polymerase || (forms messenger or MRNA)
RNA polymerase III (forms transfer or tRNA)
So, it is clear that for transcription RNA
polymerase II is required. This RNA
polymerase unwinds the two templates of the DNA.
From the figure given below, it is very clear that
new mRNA is formed and
nucleotide sequence of newly formed MRNA is exactly
same as of the nucleotide sequence of sense strand (which contains biological
information) with a difference of presence of uracil in spite of thymine.
Requirements for transcription:
(1) DNA template
(ii) |The enzyme like RNA polymerase.
(iii) All four types of ribonucleosides
triphosphates (ATP, CTP, GTP and UTP), are required because these ribose
nucleotides join to form RNA chain.
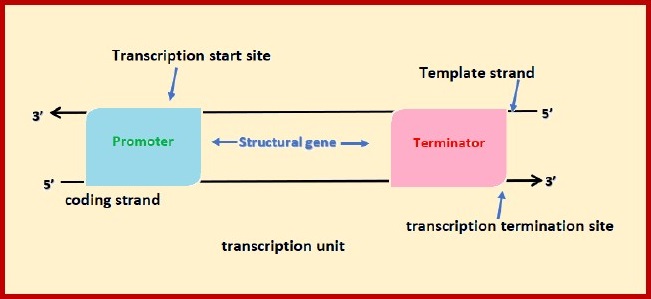
Transcription unit:
Transcription unit is that segment of DNA which is
participating in transcription.
This unit in case of prokaryotes is polycistronic
(having information for many
cistrons or proteins).
In case of eukaryotes it is mono-cistronic (having
information for one cistrons or proteins).
Transcription unit of DNA molecule
comprises of three regions-
(i) A promoter
(ii) The structural gene
(iii)A terminator
The promoter is small sequence of DNA which provides
binding site for RNA polymerase and is located towards the 5’ end of the
structural gene. It is also called the upstream end.
The structural gene is the gene to be transcribed.
The two strands of the DNA in the structural gene
have opposite polarity, i.e.one strand has 3’ — 5’ polarity and other strand
has 5’ — 3’ polarity.
Strand having 3’ — 5’ polarity acts as a template
and referred as template
strand or anticoding strand.
Strand with 5’ — 3’ polarity acts as a coding strand
or sense strand.
All the biological information is present on the
coding or sense strand.
The terminator is small sequence of DNA which
provides binding site for
terminator factor and is located towards the 3’ end
of the structural gene and
also called as downstream end.
The promoter part is A (adenine), T (thymine) rich
base area and also known
as TATA box.
It was discovered by Pribnow and also called Pribnow
box.
The terminator part is having palindromic sequence
or poly A rich base
area.
Stages of transcription: There are three stages of
transcription:
(i) Initiation
(ii) | Elongation
(iii) Termination
Initiation:
The RNA polymerase enzyme binds to the promoter
region with the help of sigma (o) factor in case of prokaryotes and with transcription
factors in eukaryotes.
Sigma (0) factor or Transcription factor and RNA
polymerase together,
forming a transcription initiation complex.
Elongation:
RNA polymerase moves along the DNA and causes
unwinding and splitting of DNA strands.
The area where DNA strand unwinds itself, become
bulgy and known as transcription bubble.
This area gives site for the formation of RNA.
Now RNA polymerase will start moving from the
promoter to the structural gene and will start forming RNA in the direction of
5’ — 3’ over the template strand which have the sequence of 3’ — 5’.
Now the ribonucleotides start coming over the
template strand and start pairing with the bases on the template DNA strand
with the help of hydrogen bonds.
The RNA polymerase helps in unwinding of DNA strand
and in polymerization of ribonucleotides by forming phosphodiester bond, to
form long chain of RNA in the 5’ -3’ sequence.
RNA polymerase utilizes ribonucleotide triphosphates
(ATP, GTP, CTP and
UTP) for the formation of RNA by the formation of
phosphodiester bonds.
Termination:
When RNA polymerase enzyme reaches the terminator
area, then the rho (po) factor of RNA polymerase enzyme separates the RNA
polymerase enzyme and newly formed mRNA from the terminator area in
prokaryotes.
RNA synthesized from 5’ end to 3’ end, antiparallel
to template strand.
Now the RNA formed will be processed further.
Many non-coding areas (introns) are also formed on
new MRNA.
Now non coding areas (introns) from newly formed RNA
are removed by
enzymes called nucleases (splicing).
Coding areas (exons) are re-joined by ligase enzyme
(union) to form a long
chain of mature MRNA.
At 5’ end a 7-methyl guanosine cap is added.
At 3' end a poly adenosine (poly A) tail is added to
form a complete mRNA.
Let us know what we have learnt!
PART: A VERY SHORT ANSWER TYPE
QUESTIONS:
1. Multiple choice questions
(i) The RNA formed after transcription is:
a) mRNA
b) rRNA
c) tRNA
d) All the above
(ii) The process of transcription occurs in
what phase of interphase of cell cycle.
a) G; phase
b) G2 phase
c) S phase
d) Gi and G2 phase
(iii) Where the process of protein
synthesis occurs in a cell?
a) in nucleus
b) in cytoplasm
c) inside ribosomes
d) All the above
(iv) What is the correct sequence of the stages
of transcription?
a) Initiation, Elongation, Termination
b) Termination, Elongation, Initiation
c) Elongation, Initiation, Termination
d) Initiation, Termination, Elongation
(v) Which RNA polymerase is required in
translation?
a) RNA polymerase |
b) RNA polymerase II
c) RNA polymerase III
d) All the above
2. Fill in the blanks
(1) In transcription, elongation stage isfollowedby
(2) The area where DNA strand unwinds itself is
known as ,
(3) TATA box of promoter in transcription unit is
also known as ;
3. True / False
(1) In prokaryotes, rho (p) factor of RNA polymerase
enzyme helps in initiation in
transcription.
(2) The promoter site is located towards the 3’ end
of the structural gene.
(3) DNA can not come out from nucleus to synthesise
the proteins.
ANSWER KEY: PART -A
(i) (a) MRNA
(ii) (d) G; and G2 phase
(iit) (b) in cytoplasm
(iv) (a) Initiation, Elongation, Termination
(v) (b) RNA polymerase II
(1) Termination
(2) Transcription bubble (because this area of DNA
strand becomes bulgy due to splitting of two strands of DNA and a bubble-like
structure appears there)
(3) Pribnow box because It was discovered by
Pribnow.
(1) False, because for transcription the initiation
factor is sigma.
(2) False, the promoter site is located towards the
5’ end of the structural gene.
(3) True, because of its larger size, DNA can not
come outside the nucleus.
PART: B SHORT ANSWER TYPE QUESTIONS
(1) Why transcription is required?
(2) Name the three types of RNA polymerase. What
function they perform?
(3) What do you mean by polycistronic unit of
transcription?
PART: C LONG ANSWER TYPE QUESTIONS
(1) What do you understand by transcription. Discuss with the help of steps
or stage involved in transcription.
A61
INTRODUCTION
Students, as discussed in previous topics that
Central Dogma is the process
in which DNA is decoded by RNA to makes proteins.
The process by which DNA is
copied to RNA is called transcription, and that by
which RNA is used to produce
proteins is called translation. In our today’s topic
we will discuss Translation in detail.Translation is the process of translating
the sequence of MRNA (messenger RNA)into a sequence of amino acids. This
translation takes place during protein
synthesis.
This process involves the transport of amino acids
to the ribosome, where they are
assembled in the polypeptide chain. After which they
will assemble into proteins
somewhere in the cytoplasm. This process is
accomplished by ENA and occurs in several stages.
THE PROTEIN SYNTHESIZING MACHINERY:
The key components required for
translation are:
Ribosomes:
The ribosome is a complex organelle, present in the cytoplasm,
which serves as the site of action for protein
synthesis. It provides the
enzymes needed for peptide bond formation.
mRNA: mRNA is used to convey information from DNA to
the ribosome. It is a single strand molecule, complimentary to the DNA
template, and is generated through transcription. Strands of MRNA are made up
of codons,each of which signifies a particular amino acid to be added to the
polypeptide in a certain order.
RNA: This is a
single strand of RNA composed of approximately 80ribonucleotides. Specific t
RNAs binds to sequence on the MRNA template and
add the corresponding amino acid to the polypeptide
chain. Therefore, t RNAs
are the molecules that actually “translate” the
language of m RNA into the language of Proteins.
RNAs need to interact with three
factors:
They must be recognized by the correct Aminoacyl
Synthetase.
They must be recognized by ribosomes.
They must bind to the correct sequence in MRNA.
Aminoacyl tRNA synthetase: Through the process of t
RNA “charging,”each t RNA molecule is linked to its correct amino acid by a
group of enzymes called Aminoacyl t RNA synthetases.
At least one type of Aminoacyl t RNA synthetase
exists for each of the 20
amino acids.
MECHANISM OF PROTEIN SYNTHESIS:
Protein biosynthesis involves following
major steps:
1. Activation of amino acids:
Here, when an Amino Acid (AA) and Adenosine Triphosphate (ATP), is mediated by
Aminoacyl ynthetases, enzyme in the presence of Mg? the amino acid- enzyme -AMP
complex is formed. The
complex remains temporarily associated with the
enzyme. The amino acid-AMP-enzyme complex is called an activated Amino Acid.
2. Charging of tRNA:
The enzyme complex formed above is reacted withthe specific t RNA. As a result,
an Amino Acid is transferred into t RNA and the enzyme and AMP are liberated.
The reaction is catalyzed by the same Aminoacyl-t RNA Synthetase enzyme. The
resulting tRNA-amino acid complex is called a charged tRNA.
3. Initiation of polypeptide chain:
Now the charged t RNA shifts to the ribosome. The large and small are the two
subunits of the ribosome. The MRNA binds to the small subunit and the charged t
RNA bind to the larger subunit to complete the initiation complex.
The Aminoacyl t RNA binding site (A site) and
Peptidyl Site (P site) are the two
sites of binding in large sub-units.
The mRNA chain has at its 5’ end an “initiator” or “start” codon (AUG) that signals
the start of polypeptide formation or
Translation.
This start codon lies close to the P site of the
ribosome.
The amino acid Formyl-Methionine (methionine in
eukaryotes) initiates the
process. It is carried by t RNA having UAC anticodon
which bonds to AUG initiator
codon of MRNA by hydrogen bonds.Initiation factors
(IF 1, IF 2 and IF 3) and GTP promote the initiation process.
4. Elongation:The first codon on MRNA binds with the anticodon of the methionyl t RNA complex in the P site.
The other Aminoacyl t RNA complex with the appropriate amino acids
thus enters the ribosome and attaches to A site.The peptide bond is formed
between the first and second amino acids, when the anticodon of new t RNA binds
to the second codon in the m RNA in the presence of an enzyme, Peptidyl Transferase.Then,
the translocation takes place i.e. when the first amino acid and the tRNA bond
broken and both get released out from the complex from E site.The second t RNA
from the A-site is pulled to the P site along with the m RNA.
5. Termination:Two
conditions are necessary for termination of protein synthesis.
One is the presence of a stop codon that signals the
chain elongation to terminate,
and the other is the presence of release factors
(RF) which recognise the chain
terminating signal.There are three terminating
codons, UAA, UGA and UAG for which t RNAs do not exist. Termination of
polypeptide chain is signaled by one of these codons in the MRNA. Release of
the Peptidyl t RNA from the ribosome is promoted by three
specific release factors, RF 1, RF2 and RF3.RF1
recognizes triplets UAA and UAG, while RF2 recognizes UAA and UGA. The
third factor RF3 does not possess any release
activity of its own, but it stimulates the binding of RF1 and RF2 with the
ribosome.
LET US KNOW WHAT WE HAVE LEARNT
PART: A VERY SHORT ANSWER TYPE
QUESTIONS:
A) MCQ’S
1. Translation is the process of
polymerisation of -
(a) amino acids
(b) sugars
(c) fats
(d) energy units
2. Which of the following is responsible
for the initiation of RNA polymerase activity?
a) initiation site
b) promoter region
c) sigma factor
d) rho factor
3). Translation occurs ina .....
(a) nucleus.
(b) cytoplasm
(c) ribosome
(d) lysosome
4). Which is energy rich molecule required
for the initiation of translation....
(a) ATP
(b) GTP
(c) CTP
(d) AMP
5). Which of the following step of
translation does not consume a high
energy phosphate bond.
(a) Translocation
(b) Amino acid activation
(c) Peptidyl transferase reaction
(d) Aminoacy] tRNA binding to A-site
B) FILL Ups:
1) A translational unit in mRNA is the sequence of
RNA that is flanked by the
start codon, ........., and the codes for a
polypeptide.
2) The cellular structure responsible for
synthesizing roteinsis............
C) TRUE/ FALSE
1. Translation takes place before transcription.
2. E.coli bacteria can synthesize all of the amino
acids required for protein
synthesis.
ANSWERKEY: PART-A
A)MCQS
1. (b) amino acids _ Translation is ail about
protein synthesis. Polymer of
Amino acids joined together by peptide bond to form polypeptides.
2. (c) sigma factor is a protein needed for
initiation of RNA polymerase
activity.
3. (c) Ribosome — is the structural unit in cell
where protein synthesis takes place
4. (b) GTP — is required for translation initiation,
for elongation and for
termination.
5. (c) Peptidyl transferase reaction involves
formation of peptide bond.
B) FILL UPS:
(a) stop codon
(b) Ribosome
C) TRUE/FALSE:
(a) FALSE; According to Central Dogma first step is
DNA replication then
transcription and the last step is translation.
(b) TRUE
PART: B SHORT ANSWER TYPE QUESTIONS:
Q1) What is the function of mRNA?
Q2) Differentiate between transcription and
translation?
Q3) What do you mean by translocation?
PART: C LONG ANSWER TYPE QUESTIONS:
Q1) Describe the process of Translation?
A62
INTRODUCTION
The DNA is a blue print of life. It carries vital
information of life but how information get translated into protein is still a
mystery which has kept the
scientist waiting for a long time. This mystery was unfolded
when the GENETIC
CODE was understood by the scientist in the field of
genetics. James Watson and Crick proposed double helix modal for structure of
DNA and got Nobel Prize.
During Replication and Transcription, a nucleic acid
was copied to form another nucleic acid. The process of translation requires
transfer of genetic information from a polymer of nucleotides to synthesise a
polymer of amino acids. Change in nucleic acids were responsible for change in
amino acids in proteins. It was very challenging for scientists to determine
the biochemical
nature of genetic material and the structure of DNA.
They found it exciting
deciphering of genetic code. This was a complex
process. Many scientists
were involved from different disciplines.
George Gamow, a physicists, who came with the idea
that there are only 4 nucleotide bases and they code for 20 amino acids.
It means the code should be a combination of bases.
The combination of 4° i.e. (4 x 4 x 4) would
generate 64 codons, many more codons than required.
It was hard task to provide proof that the codon was
a TRIPLET.
Indian origin scientist Har Gobind Khorana developed
chemical method that was helpful in synthesising RNA molecules with defined
combination of bases.( homopolymers and copolymers)
A Homopolymer is made up of only one type of
monomers and a Copolymer is made of two or more monomers.
Marshall Nirenberg and Heinrich J. Matthaei were the
first to discover the nature of codon.
At first polyuracil RNA sequence UUU coded for
Phenylalanine.
The Severo Ochoa enzyme was discovered by scientist
Severo Ochoa.
It is Polynucleotide Phosphorylase (PNPase) which is
bifunctional enzyme with phosphorolytic 3° to 5’ exoribonuclease activity and
3’ terminal oligonucleotide polymerase activity.
The function of this enzyme is to dismantie the long
RNA chain that start from 3' end and move toward 5’ end.
Finally the checker board for genetic code was
prepared.
GENETIC CODE:Genetic
code is the sequence of nucleotides that codes for the corresponding amino acid
sequence of proteins.
1) Itis TRIPLET. Out of 64 codons, 61 codons code
for amino acids and 03
codons do not code for any amino acid.
2) One codon codes for only one amino acid, hence it
is unambigous and specific.
3) Some amino acids are coded by more than one
codon. Hence the code is degenrate.
4) The codon is read in MRNA in contiguous fashion
there are no punctuations.
5) The code is nearly universal; for example. From
bacteria to human UUU
would code for phenylalanine (phe). Some expections
to this rule have been
found in mitochondrial codons and in some
Protozoans.
6) AUG has dual function it codes for methionine and
it also act as initiator code.
7) 61 Sense codon and 03 Nonsense codons are
there.UAA, UAG, UGA are STOP CODONS or NON- SENSE CODONS.
MUTATIONS & GENETIC CODE
The phenomena of change occurring in DNA sequence
caused either by external
factors or internal factors like smoking and UV rays
is termed as mutation.
Variation caused by change in building block and
single base pair of DNA is termed
as POINT MUTATION example is SICKLE CELL ANAEMIA
When reading frame of genetic code is altered by insertion or deletion of one
or two bases leads to frame shift mutation.
tRNA ADAPTER MOLECULE
tRNA is also called SRNA soluble RNA, or activator
RNA.has a role as adapter
molecule. It has an anticodon loop having bases
complimentary to the code. It
has amino acid accepter. The secondary structure of
tRNA looks like CLOVER
LEAF. In actual structure tRNA looks like inverted
LET US KNOW WHAT WE HAVE LEARNT!
A) MULTIPLE CHOICE QUESTIONS:
1. Which of the following is not a feature
of genetic code?
a) Triplet
b) degenerate
c) Ambiguous
d) Non overlapping
2. The codon is a .
a) Singlet
b) Triplet
c) Doublet
d) Quadruplet
3. Who was instrumental in finding chemical
method of determining DNA
a) Severo Ochoa
b) George Gamow
c) Har Gobind Khorana
d) Marshall Nirenberg
4. Sickle cell anemia is an example of
a) point mutation
b) both of these
c) silent mutation
d) none of these
5. The secondary stucture of t RNA looks
like
a) clover leaf like structure
b) flower like structure
c) inverted L shape structure
d) inverted T shape structure
B) FILL IN THE BLANKS:
1. The codon AUG code for amino acid methionine also
serves as___.
2. tRNA has an anticodon loop that has bases
.........to the code.
C) TRUE/FALSE
1) Some amino acids are coded by more than one
codon, so the genetic code
is degenerative.
2) AUG codes for lysine.
3) Severo Ochoa enzyme is polynucleotide
phosphorylase.
A) MULTIPLE CHOICE QUESTIONS:
1. c) Ambiguous The genetic code is unambiguous and
specific.
2. b) Triplet genetic code is triplet
3. c) Har Gobind Khorana
4. a) point mutation
5. a) clover leaf like structure
B) FILL UPs:
1) Initiator codon
2) Complementary
C) TRUE/ FALSE
1) True
2) False AUG codes for Methionine and also acts as
initiator codon
3) True
PART:B SHORT ANSWER TYPE QUESTIONS:
1) Discuss role of Har Gobind Khorana in defining
Genetic code.
2) How tRNA is the adapter molecule?
3) What are frame shift mutations?
PART:C LONG ANSWER TYPE QUESTION:
1) Write the salient features of Genetic Code.
A63
INTRODUCTION
In your previous assignment, you have studied about
the genetic code. Now,you know that the genetic code is the set of rules used by
living cells to translate information encoded within the genetic material (DNA
or mRNA) into proteins.All the genes present in the chromosome are not
expressed simultaneously.
The cell permits the expression of only a few genes
at a time, thus maintaining its
economy. So, in this assignment, we will discuss
about the regulation of gene
expression.Gene expression is the mechanism at the
molecular level by which a gene is
able to express itself in the phenotype of an
organism.
OPERON CONCEPT
From a number of studies on the metabolism of the bacterium Escherichia Coli,two French scientists, Jacob and Monod proposed a model of gene regulation,known as the Operon model.
Operon is a co-ordinated
group of genes such as Structural Gene, Operator Gene, Promoter Gene and
Regulator Gene which function together and regulate a metabolic pathway as a
unit e.g.: Lac operon.
Lac operon is an inducer operon. A segment of DNA
made up of 3- structural
genes, operator gene, promoter gene and regulator
gene.
a) Transcribes mRNA for polypeptide synthesis.
b) It is the gene which receives the product of the
regulator gene. It allows the functioning of the operon when it is not covered
by the biochemical produced by the regulator gene.
c) Provides the attachment site for the RNA
polymerase
d) It synthesises a biochemical or regulator protein
which can act positively as activator and negatively as repressor. It controls
the
activity of the operator gene.
e) and or co-repressor (from outside) are also
found.
LAC OPERON- INDUCIBLE OPERON SYSTEMS
The lac refers to lactose. In E. Coli, breakdown of
lactose requires three
enzymes. These enzymes are synthesised together in a
co-ordinated manner
by functional unit of DNA, i.e., fac operon. Since
the addition of lactose itself
stimulates the production of the required enzymes,
thus it is called inducible
operon system.
The lac operon consists of the
following:
a. Three Structural Genes: Three structural genes
are:
1. Lac z: The z gene codes for B-galactosidase which
is primarily responsible
for the hydrolysis of the disaccharide, lactose into
its monomeric units
galactose and glucose.
2. Lac y: The y gene codes for the permease, which
increases the permeability of the cell to B-galactosidase.
3. Lac a: The a gene codes for the transacetylase
which can transfer acetyl
group to B-galactosidase.
b. Operator Gene: It interacts with a protein
molecule or regulator molecule, which
prevents the transcription of structural genes.
c. Promoter Gene: The gene possesses site for RNA
polymerase attachment
d. Regulator gene (i): The gene codes for a protein
known as the repressor protein, it is synthesised all the time from the
i-gene,that is why it is constitutive gene which is functional always.
SWITCHING OFF THE OPERON:The operon is switched off
when the repressor protein produced by regulatory or inhibitor gene binds to
operator gene. RNA polymerase gets blocked, so there would be no transcription.
SWITCHING ON THE OPERON:
Regulation of lac operon by repressor is referred to
as negative control or
regulation. If lactose is provided in the growth
medium of the bacteria, the lactose
is transferred into the cells through the action of
Permease. A very low level of lac
operon has to be present in the cell all the time;
otherwise lactose cannot enter the
cells. In the presence of an inducer, such as
lactose or allolactose, the repressor is
inactivated by interaction with inducer. This allows
RNA polymerase access to the
promoter and transcription proceeds.
Lac operon is under control of positive regulation
as well.
PART A: VERY SHORT ANSWER TYPE
QUESTIONS
(1) Multiple Choice Questions
1. Lac operon was proposed by:
(a) Hershey and Chase
(b) Meselson and Stahl
(c) Alec Jeffreys
(d) Francois Jacob and Jacques Monod
2. The equivalent of structural gene is :
(a) Operon
(b) Recon
(c) Muton
(d) Cistron
3. Which of the following is required as
inducer in the Lac operon?
(a) Glucose
(b) Lactose
(c) Galactose
(d) Lactose and Galactose
4. The Lac operon consists of :
(a) Four regulatory genes only
(b) One regulatory gene and 3 structural genes
(c) Two regulatory genes
(d) 3 regulatory and 3 structural genes
5. In lac operon, the gene Z codes for
which enzyme ?
(a) Transacetylase
(b) B-galactosidase
(c) Permease
(d) Repressor protein
(II) FILL-UPS
1. In bacteria, the regulation of gene expression is
usually affected through .
2. Lac operon in bacteria codes for genes
responsible for the metabolism of .
3. Promoter gene provides attachment site for
(Ill) TRUE / FALSE
1. Operator gene is the site or the gene for binding
RNA polymerase in an
operon.
2. Lac operon gets switched on in E.Coli when
lactose is present and it binds to the repressor.
(1)
Multiple choice Questions:
1. (d) Francois Jacob and Jacques Monod
2. (d) Cistron. Structural genes are continuous
stretch of DNA that regulates a particular genetic trait and Cistron also
denotes a coding sequence or segment of DNA encoding a polypeptide.
3. (b) Lactose. Lactose inactivates repressor.
4. (b) One regulatory genes and three structural
genes. The Lac ‘i’ gene is
regulatory, while the three structural genes are Lac
z, Lac y and Lac a.
5. (b) B-galactosidase. B-galactosidase hydrolyzes
lactose to galactose and glucose.
(Il) Fill Ups :
1. Operons. Gene regulation is a process in which a
cell determines which gene it will express and when
2. Lactose. Lac operon has three structural genes
required for the degradation of lactose.
3. RNA polymerase. Promoter gene marks the site at
which transcription of mRNA starts and where RNA polymerase enzyme binds
(I) True / False
1. False. Operator gene is the segment of DNA to
which a repressor binds.
2. True. Lactose binds itself to active repressor
leading to change in its structure. Operon gets transcribed and enzymes are
produced.
PART B: SHORT ANSWER TYPE QUESTIONS:
1. Study the figure given below and answer the
questions that follow:
a. Name the enzyme transcribed by Z gene
b. How does the repressor molecule get inactivated
2. Name the genes that constitute an operon
3. What is the role of Lactose in lac operon
PART C: LONG ANSWER TYPE QUESTIONS:
1. Define an operon. Explain an inducible operon.
A64
INTRODUCTION
Students, as you know, the history of the human race
has been filled with
curiosity and discovery about our abilities and
limitations. With the rapid growth
of scientific knowledge and experimental methods,
humans have begun to unravel and challenge another mystery, the discovery of
the entire genetic make-up of the human body. This endeavor, the Human Genome
Project
(HGP), has created hopes and expectations about
better health care.
HUMAN GENOME PROJECT was called a mega project.
Human genome is said to have approximately 3x10°bp and if one cost of
sequencing required is US $3 per bp then approximately cost of project will be
9 billion US dollars. The HGP was initiated in 1990 under the leadership of
American geneticist Francis Collins, with support from the U.S. Department of
Energy (DOE) and
the National Institutes of Health (NIH). The effort
was soon joined by scientists from around the world.The Human Genome Project is
an ambitious research effort aimed at
deciphering the chemical makeup of the entire human
genetic code (i.e.,the genome).
GOALS
1. Identify all the approximately 20,000-25,000genes
in human DNA.
2. Determine the sequences of the 3 billion chemical
base pairs that make up human DNA.
3. Share this information in databases.
4. Improve tools for data analysis.
5. Transfer related technologies to other sectors.
6. Address the ethical, legal, and social issues
(ELSI)that may arise from the
project.
METHODOLOGIES
The method involves two major approaches.
1. EE. This method is focussed on identifying
all the genes that are expressed as RNA.
2. . This method took the blind approach of simply sequencing the whole set of
genomes that contained all the coding and
noncoding sequence, and later assigning different
regions in the sequence with
functions.
VECTORS IN HGP
The commonly used vectors were
[BBM (Bacterial artificial chromosomes) It is based
on natural, extra-chromosomal plasmid of E. coli. These vectors can accommodate
upto 300-350 kb of foreign DNA and are also being used in genome sequencing
project.
[HM (Yeast artificial chromosomes) are used to clone
DNA fragments of more than 1Mb in size. However, While YAC can be used for
cloning and sequencing, BACs are much more stable.
FREDERICK SANGER developed a principle by which
fragments were sequenced. Sanger is also credited for developing method for
determination of amino acid sequences in proteins. These sequences were then
arranged
based on some overlapping regions present in them.
This required generations of overlapping fragments for sequencing. Alignment of
these sequences was humanly not possible. Therefore, specialised computer-based
programmes were developed. These sequences were subsequently annotated and were
assigned to each other. The sequence of chromosome 1 was completed only
in May 2008.
SALIENT FEATURES OF HUMAN GENOME
1. The human genome contains 3164.7 million
nucleotide bases.
2. The average gene consists of 3000 bases.
3. Total number of genes is estimated at 30,000.
4. The functions are unknown for over 50% of the
discovered genes.
5. Less than 2% of genome codes for proteins.
6. Repeated sequences make up very large portion of
human genome.
7. Chromosomes 1 has most genes (2968) and the Y has
the fewest (231).
8. Scientists have identified about 1.4 million
locations where single base DNA
differences (SNPs-single nucleotide polymorphism)
occurs in humans.
9. The largest gene in humans is Dystrophin.
This information promises to revolutionise the
process of finding chromosomal
locations for disease associated sequences and
tracing human history.
APPLICATIONS OF HGP
1. Identification of human genes and their
functions.
2. Understanding of polygenic disorders e.g. cancer,
hypertension, & diabetes.
3. Improvement in gene therapy.
4. Improve diagnosis of diseases.
5. Development of pharmacogenesis.
6. Improved knowledge on mutations.
7. Development of biotechnology.
FUTURE CHALLENGES
We still do not know about the following: -
Gene number, exact locations and functions.
Gene regulation, DNA sequence organization
Chromosomal structure and organization and so on.
PART-A VERY SHORT ANSWER TYPE QUESTIONS
MCQ's:-
1. The Human Genome Project was initiated
by:
a) NIH and DOE
b) NIH and EBI
c) NIH and DDBJ
d) DOE and DDBJ
2. Which of the following vectors were used
in HGP?
a) Plasmid and cosmid
b) A Phage and M13 vector
c) Phagmid and shuttle vectors
d) BAC and YAC
3. According to HGP human genome consists
of:
a) 100000 genes
b) 50000 genes
c) 30000 genes
d) 20000 genes
4. YAC means:
a) Yellow artificial chromosomes
b) Yeast artificial chromosomes
c) Yeast acquired chromosomes
d) Yeast artificial chromatids
5. The largest gene in humans is:
a) Titin
b) Dystrophin
c) Insulin
d) Phosphofructokinase
FILL UP’s:-
1. The human genome contains ............ million
nucleotide bases.
2. The functions are unknown for over .....% of the
discovered genes.
TRUE /FALSE:-
1. EXPRESSED SEQUENCE TAGS method is focussed on
identifying all the
genes that are expressed as Proteins.
2. SNP means Single Nucleotide Polymorphism.
ANSWER KEY
MCQ’s:-
1. a (NIH and DOE) HGP was initiated by U.S.
Department of Energy (DOE) and the National Institutes of Health (NIH).
2. d (BAC and YAC) Both BAC and YAC were used in
HGP. While YAC can be used for cloning and sequencing, BACs are much more
stable.
3. c (30000 genes)
4. b (Yeast artificial chromosomes) used to clone
DNA fragments of more than 1Mb in size.
5. b (Dystrophin)
FILL UP’s:-
1. 3164.7
2. 50
TRUE/ FALSE:-
1. FALSE EXPRESSED SEQUENCE TAGS method is focussed
on identifying all the genes that are expressed as RNA.
2. TRUE
PART-B SHORT ANSWER TYPE QUESTIONS
1. Why is human genome project called a mega
project?
2. What are the benefits of the human genome
project?
PART-C LONG ANSWER TYPE QUESTIONS
1. Write salient features of human genome project?
A65
INTRODUCTION
DNA FINGERPRINTING
DNA fingerprinting, also called DNA typing, DNA
profiling, genetic fingerprinting, genotyping, or identity testing, in
genetics, method ofisolating and identifying variable elements within the
base-pair sequence
of DNA (deoxyribonucleic acid). It is a technique of
determining nucleotide
sequences of certain areas of DNA which are unique
to each individual. Each person has unique DNA fingerprint.
BACKGROUND:
ALEC JEFFREY'S (1984) invented the DNA finger
printing technique at Leicester University, United Kingdom.
Dr V. k. Kashyap and Dr Lal Ji Singh started DNA
fingerprinting technology,in India at CCMB (Centre for cell and molecular
biology) Hyderabad.
PRINCIPLE OF DNA FINGERPRINTING:
Human genome possesses numerous small noncoding but
inheritable sequences of bases which are repeated many times.
The area with same sequence of bases repeated
several times is called REPETITIVE DNA.
They can be separated as satellite from the bulk DNA
during density gradient
centrifugation and hence called SATELLITE DNA.
SATELLITE DNA is of two types
1. MICRO SATELLITE
2. MINI SATELLITE
Microsatellite also Known as Simple sequence repeats
(SSRs) or short tandem repeats (STRs) are repeating sequence of 2-6 base pairs
of DNA.
Mini satellite also known as (VNTR) is a section of
DNA that consist of short series of bases of 10-16 base pairs.
Their analysis is useful in genetic and Biology
research, forensic and DNA
fingerprinting.
An inheritable mutation occurring in a population at
high frequency is referred to as DNA POLYMORPHISM.
Repeated nucleotides sequence in a non-coding DNA of
an Individual is called VARIABLE NUMBER OF TANDEM REPEATS (VNTR).
STEPS USED IN DNA FINGERPRINTING
1. ISOLATION OF DNA:
The DNA is extracted from the nuclei of white blood cells, blood, semen, hair
follicle cells.
2. DNA AMPLIFICATION:
The extracted DNA is amplified through PCR (Polymerase chain reaction) It
produces numerous copies of DNA.
3. FRAGMENTATION: The DNA molecule
are first broken with the help of enzyme RESTRICTION ENDONUCLEASE (called
Chemical knife) that cuts them into fragments.
4. SEPARATION OF VNTRs CHOPPED:
DNA is exposed to electrophoresis over agarose polymer gel. It separates DNA
fragments. The separated VNTRS can be recognized by staining them Acridine dye
that become fluorescent
when illuminated with UV Radiations.
5. SINGLE STRANDARD DNA:
VNTRS are treated with alkaline chemicals to split them in to single stranded
DNAs.
6. SOUTHERN BLOTTING: The separated
VNTR single stranded sequences are transferred to nylon membrane placed over a
gel. The procedure is called SOUTHERN BLOTTING.
7. DNA PROBES:
They are small radioactive single stranded DNA segments of known sequences of
nitrogen bases.
8. HYBRIDISATION:
Nylon sheet is immersed in a bath to which DNA probes are added. The probes get
attached to single stranded VNTRs_ having complementary nucleotide sequence.
Rest of probes are worked away.
9. EXPOSURE TO X-RAY FILMS: The nylon
membrane containing the radioactive DNA probes and VNTRs is exposed to X-Ray
film. The hybridized radioactive VNTRs appear as dark bands. The film gives us
DNA prints. The
prints are compared and relationship found.
APPLICATION OF DNA FINGER PRINTING
1. INDENTIFICATION:
DNA fingerprinting is sure method of identification of criminals involved in
various type of crimes including rape and murder.
2. PATERNITY- MATERNITY DISPUTES:
The method can provide reliable information as to real biological father,
mother on off spring.
3. HUMAN LINEAGE: It provides
information to human lineage and relationship with apes.
4. MIGRATIONS: DNA finger
printing can identify racial groups,
origin, historical migration and invasions.
5. DNA
fingerprinting is used to identify and protect the commercial varieties of
crops and livestock.
LET US KNOW WHAT WE HAVE LEARNT
PART: (A) VERY SHORT ANSWER TYPE
QUESTIOS:
(i) Multiple Choice Questions:
1. DNA finger-printing was developed by:
(a) Francis Crick
(b) Knorana
(c) Alec Jeffery
(d) James Watson
2. The technique to distinguish the
individuals based on their DNA print
patterns is called:
(a) DNA fingerprinting
(b) DNA profiling
(c) Molecular fingerprinting
(d) All of these
3. Which tissue samples are used for DNA
fingerprinting?
(a) Hair
(b) Skin
(c) Blood
(d) Any of above
4. Which one is used in DNA
Finger-printing?
(a) Western Blotting
(b) Flow Cytometry
(c) Northern Blotting
(d) Southern Blotting
5. DNA Fingerprinting is applied in:
(a) Identification of criminals
(b) Paternity disputes
(c) Identify and protect Commercial varieties of
plants
(d) All of these
(ii) Fill ups:-
6. Polymorphism arises due to .
7. DNA molecules are broken with the help of an
enzyme known as
8. Mini-satellites are tt.
(iii) True/ False:
9. The DNA finger pattern of child is 100% similar
to the father’s DNA print.
10. The sequences of satellite DNA do not code for
proteins.
11. DNA can be used as a useful tool in the
forensics application by the presence of lysozymes in it.
A) Multiple Choice Questions:
1. (c) ALEC JEFFREY, he developed this technique in
1984
2. (d) All of the above DNA fingerprinting, also
called DNA typing, DNA profiling, genetic fingerprinting, genotyping, or
identity testing is the method of isolating and identifying variable elements
within the base-pair sequence of DNA (deoxyribonucleic acid).
3. (d) All of the above DNA Sample can be obtained
from any tissue and can be
amplified by PCR.
4. (d) Southern Blotting It is the process of
separation of VNTRs from agrosegel to nylon membrane.
5. (d) All of these
B) Fill-Ups:
6) Mutation — Polymorphism arises due to mutation.
It can also be known as the
discontinuous variation that is seen among the
individuals of species.
7) Restriction Endonuclease - It is also called as
chemical knife that cuts DNA
molecule in to fragments.
8) Minisatellites - They are the SHORT NON CODING
REPITITIVE SEQUENCES present throughout the chromosomes.
C) True/False:
9) False; The DNA fingerprint pattern of a child is
50% bands similar to the father and
rest 50% to the mother.
10) True ; The small peaks formed by the DNA when
kept in the density gradient
centrifugation are called as SATELLITE DNA. These
DNA sequences do not code for proteins. On the contrary, they form a large
portion in the genes of humans
11) False; DNA can be used as a useful tool in the
forensics application by showing
some degree of polymorphism with hair follicles.
PART: (B) SHORT-ANSWER TYPE QUESTIONS:-
1. Whatis the principle of DNA fingerprinting?
2. What do you mean by satellite DNA?
PART: (C) LONG-ANSWER TYPE QUESTIONS:-
1. What is DNA fingerprinting? Explain the different
steps used in DNA fingerprinting with a suitable diagram.
A66
Hello students, we have discussed all the topics of
Molecular basis of inheritance. Let us work on the NCERT questions of the
chapter. Now it is time to test our understanding of the content and we should
be able to answer the related questions.
RECAPITULATION
DNA and RNA are the two types of nucleic acids found
in organisms.
DNA is a long polymer of deoxyribonucleotides.
In RNA every nucleotide residue has an additional
-OH group present at 2'position of ribose.
Double helix of DNA is made of two nucleotide
chains.
Euchromatin is transcriptionally active chromatin,
whereas Heterochromatin
is inactive
The human genome contains 3164.7 million nucleotide
bases.“Polymorphism variation of genetic level arises, due to mutation.
NCERT EXERCISE QUESTIONS [1 to 14]
Question 1Group the following as
nitrogenous bases and nucleosides:Adenine, Cytidine, Thymine, Guanosine, Uracil
and Cytosine.
Answer 1Adenine, Uracil, Cytosine, Thymine are
nitrogenous bases and Cytidine
Guanosine are nucleosides.
Question. 2If a double stranded DNA has 20
percent of CYTOSINE, calculate the percentage of ADENINE in the DNA.
Answer 2According to Chargaff’s rule —
A+G=C+T=1
More over AMOUNT of ADENINE =AMOUNT of THYAMINE.And
AMOUNT OF GUANINE =AMOUNT OF CYTOSINE.Since percentage of CYTOSINE is 20%.
Percentage of GUANINE is also 20%.Now we are left with 60%. This 60% will be
half Adenine and half Thymine i.e.30% Adenine and 30% Thymine.
Question.3If the sequence of one strand of
DNA is written as follows:
5' -ATGCATGCATGCATGCATGCATGCATGC-3'
Write down the sequence of COMPLEMENTARY
STRAND in 5'—3' direction.
Answer 3
5 GCAT GCAT GCAT GCAT GCAT GCAT GCAT 3
The two strands of DNA have antiparallel polarity.
Question.4If the sequence of the coding strand in a
transcription unit is written as
follows: 5' -ATGCATGCATGCATGCATGCATGCATGC-3' Write
down the sequence of mRNA.
Answer 4
The sequence of m RNA shall be :
5-UACGUACGUACGUACGUACGUACGUACG-3
Question.5Which property of DNA double
helix led Watson and Crick to hypothesis semi-conservative mode of DNA
replication? Explain.
Answer 5The semi conservative mode of DNA
replication hypothesised by Watson
Crick was based upon the property that during replication the Two strands would separate and act as a template for the synthesis of new complimentary
strands. After the completion of replication,
each DNA molecule would have one parental and one newly synthesised strand.
This scheme was termed as
“Semi conservative DNA replication.
Question 6.Depending upon the chemical
nature of the template (DNA or RNA)and the nature of nucleic acids synthesised
from it (DNA or RNA), listthe types of nucleic acid polymerases.
Answer 6Process of replication needs a set of
enzymes. Depending upon the chemical
nature of the template( DNA or RNA) and the nature
of the nucleic acids synthesised from it, there are four different types of
nucleic acid polymerases:
(i) DNA-dependent DNA polymerase - Uses DNA template
to catalyse the polymerisation of deoxynucleotides.
(ii) DNA-dependent RNA polymerase- Catalyses
transcription of all types of RNAs in bacteria.
(iii) In eukaryotes there are three types of DNA-
dependent RNA polymerase:
(a) RNA polymerase I- Transcribes rRNAS
(b) RNA polymerase Il- Transcribes precursor of MRNA
i.e. hnDNA
(c) RNA polymerase Ill-Transcribes tRNA, srRNA and
snRNAS. .
(iv) Reverse transcriptase for the synthesis of
complementary DNA over RNA template.
Question.7How did Hershey and Chase
differentiate between DNA and protein in their experiment while proving that
DNA is the genetic material?
Answer 7 Harshey and Chase (1952) worked on viruses
which infect bacteria called
bacteriophages .They concluded with their experiment
that bacteria which were infected with viruses that had radioactive DNA were
found to be radioactive.It gave the idea that DNA acted as hereditary material
which was
transmitted from bacteriophage to bacteria.Bacteria which was infected with
bacteriophages (VIRUSES) that has radioactive
proteins. They were not found to be radioactive. It indicates that proteins are
not transported to bacteria.
Question.8Differentiate between the
followings:
(A) Repetitive DNA and Satellite DNA
(B) MRNA and tRNA
(C) Template strand and Coding strand
Answer 8. (A)Repetitive DNA refers to the sequences
of DNA when a small stretch of DNA
is repeated many times. They code for
proteins.Satellite DNA refers to those repetitive DNA sequence which do not
code for
any protein but form a large protein of the gene.
(B)mRNA It is the straight chain molecule. Triplets
of Nitrogenous bases (in the
form of codons) code for the specific amino acids
and decides the sequence of
amino acid in a polypeptide. It has highly
methylated 5’ cap Poly A (a long chain of adenine nucleotides) 3° tail.tRNA
This molecule becomes a looped and assumes a clover leaf like structure. It has
4 important binding sites. At one end is a sequence of three bases called
anticodon, opposite to it is amino acid arm, another arm is ribosome binding
site and finally enzyme binding site.
(C)Template strand this is the strand of DNA with 3°
> 5 polarity that functions
as template for transcriptions.Coding strand this is
the strand of DNA with 5 > 3° polarity that does not code for region of RNA
Question.9Enlist two essential roles of
ribosome during translation.
Answer 9) Role of ribosomes
1) Ribosomes usually forms linear or helical groups
during the active protein
synthesis called polyribosomes or polysomes .
2) Ribosomes acts as a catalyst for formation of
peptide bond by an enzyme peptidyl transferase .
Question.10In the medium where E. coli was
growing, lactose was added, which induced the lac operon. Then, why does lac
operon shut down some time after addition of lactose in the medium?
Answer.10The addition of lactose induced the lack
operon. As lactose formed inducer —
repression complex, the operator gene got switched
on. As the lactose were digested by the action of enzymes repressor became free
and operator gene gets switched off.
Question.11Explain (in one or two lines)
the function of the followings:
(A) Promoter
(B) tRNA
(C) Exons
Answer 11(A)Promoter :It is the region of DNA where
RNA polymerase attaches prior to the
process of transcription
(B)tRNA :It works as adaptor molecules for carrying amino
acids to the mRNA template during protein synthesis .lt bears anticodon and
recognises the specific codon on MRNA .
(C)Exons: The region of a gene, which become part of
m RNA and code for different regions of the protein are referred to exons
Question.12Why the Human Genome project is
called a mega project?
Answer 12.Human Genome Project is called a
Mega Project due to following reasons:
1. HGP took approximately 13 years
to accomplish in 2006.
2. Human genome is set to bear 3x10° bp. The cost of
the product can be imagined that if the cost of sequencing a base pair is $3
sequencing of {3x10}° base pair cost comes out to be
9 billion US dollars.
3. If such sequences are stored in books, with every
page having 1000 letters and each book is of 1000 pages, total number of books
formed will be 3300 to store information present in single human cell thus for
storing this data high speed computers are required.
4. Because of this project, many new areas have
opened in genetics.Depending on the high magnitude and requirements of the
project, HGP has been called mega project.
Question.13What is DNA fingerprinting?
Mention its application.
Answer 13 DNA fingerprinting, also called DNA
typing, DNA profiling, genetic
fingerprinting, genotyping, or identity testing, in
genetics, method of isolating
and identifying variable elements within the
base-pair sequence of DNA (deoxyribonucleic acid).DNA fingerprinting technique
was developed by Alec Jeffery (1985.86) at
Leicester University, United Kingdom. Inheritance of
DNA is very stable, every
person has specific pattern of sequence which shows
a combination of DNA of both mother and father.Applications of DNA
fingerprinting ;
1) Paternity disputes can be solved by DNA
fingerprinting
2) It can solve the problems of evolution
3) It can be used to study the breeding patterns of
animals facing the danger
of extinction
4) It is useful in restoring health of the patients
suffering from the leukemia
(blood cancer )
5) It is very useful in the detection of crime and
legal persuits
Question.14 Briefly describe the following:
(a) Transcription
(b) Polymorphism
(c) Translation
(d) Bioinformatics
Answer 14(a)Transcription
It is one of the fundamental processes that happens
to our genome. It's the process of turning DNA into RNA. And you may have heard
about the central dogma, which is DNA, to RNA, to protein. Well, transcription
refers to that first part of going from DNA to RNA. And we transcribe DNA to
RNA in specific
places. The most popular places are those things
that code for these protein-
encoding genes. But there are a whole host of other
RNAs that get transcribed, like transfer RNAs and ribosomal RNAs, that do other
functions that are genomic as well.
(b) Polymorphism As in DNA sequence is the basis of
genetic mapping of human genome as
well as of DNA fingerprinting, it is essential that
we understand what DNA
polymorphism means in simple terms. A DNA
Polymorphism is a sequence difference compared to a reference standard that is
present in a least 1-2% of a population. Polymorphism (variation at genetic
level) arises due to mutations. New mutations may arise in an individual either
in somatic cells or in the germ cells.
(c) Translation is the process in which ribosomes in
the cytoplasm or endoplasmic reticulum synthesize proteins after the process of
transcription of DNA to RNA in the cell's nucleus. The entire process is called
gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome,
outside the nucleus, to produce a specific amino
acid chain, or polypeptide.
The polypeptide later folds into an active protein
and performs its functions in
the cell. The ribosome facilitates decoding by
inducing the binding of complementary tRNA anticodon sequences to mRNA codons.
The tRNAs
carry specific amino acids that are chained together
into a polypeptide as the
mRNA passes through and is "read” by the
ribosome.
(d) Bioinformatics is an interdisciplinary field
that develops methods and software tools for understanding biological data, in
particular when the data sets are large and complex. As an interdisciplinary
field of science,bioinformatics combines biology, computer science, information
engineering,
mathematics and statistics to analyse and interpret
the biological data.Bioinformatics has been used for in silico analyses of
biological queries using mathematical and statistical techniques.
A67
INTRODUCTION
Dear students, we have got in depth knowledge of
Genetic material in this chapter. We have got knowledge about strucure of DNA
and why DNA is suitable to be genetic material. We have gone through DNA
replication,
Transcription and Translation. We read about Human
Genome Project and DNA Fingerprinting. We will revise the chapter by learning
important Definitions and Differences related to the content we have studied.
IMPORTANT DEFINITIONS NUCLEOTIDE DNA
and RNAi are polymers of deoxyribonucleotides and
ribonucleotides. A nucleotide has three components —
1) A nitrogenous base
2) A deoxyribose pentose sugar (DNA) OR ribose
pentose sugar (RNA)
3) A phosphate group
There are two types of nitrogenous bases-
Purines ( Adenine and Guanine)Pyrimidines ( Cytosine
, Uracil, Thymine)We will be able to learn about the differences between
Purines and Pyrimidines. Both have a pyrimidine ring ( similar to Benzene ring
) . But purines have pyrimidine ring
fused to an imidazole ring.
When a nirogenous bases are linked to pentose sugar
through N- glycosidic bond, there is formation of a Nucleoside. The phosphate
group is linked through a phosphoester linkage to Nucleoside to make it a
Nucleotide.
REPLICATION, TRANSCRIPTION AND
TRANSLATION
Replication is when DNA synthesis takes
place.Transcription is when genetic information is copied from DNA to RNA.
Translation is process of polymerisation of amino
acids to form polypeptide inturn
leading to protein synthesis. The order and sequence
of amino acids are defined by
the sequence of bases in the MRNA.
Francis Crick proposed central dogma in molecular
biology. It states that the
genetic information flows in a specific sequence —
DNA ---—----—- mRNA --—--------------—- Protein
NUCLEOSOME
When the negatively charged DNA is wrapped around
the positively charged
histone octamer , it becomes a Nucleosome.
OKAZAKI FRAGMENTS:
Okazaki fragments are short sequences of DNA
nucleotides with approimately 150-200 base pairs which are synthesised on the
lagging replication. The ligase enzyme joins the Okazaki fragments to make one
strand during DNA replication.
GENETIC CODE:Genetic
code is required for TRANSLATION.Genetic code is the set of rules ( triplet of
base pairs) used to translate
information encoded within genetic material to
specify particular amino acids to make proteins.
LET US KNOW WHAT WE HAVE LEARNT!
PART:A VERY SHORT ANSWER TYPE
QUESTIONS:
A)MULTIPLE CHOICE QUESTIONS:
1) In a nucleotide nitrogenous base is
linked to pentose sugar through —
(a) N-glycosidic bond
(b) phosphoester bond
(c) hydrogen bond
(d) hydroxyl bond
2) Replication refers to —
(a) Protein synthesis
(b) RNA synthesis
(c) DNA synthesis
(d) none of these
3) Genetic code is required in the process
of
(a) Replication
(b) Translation
(c) Transcription
(d) All of these
4) The process of copying genetic
information from one strand of DNA into RNA is termed as-
(a) Translation
(b) Transcription
(c) Replication
(d) none of these
5) The negatively charged DNA is wrapped
around positively charged histone octamer to form —
(a) Nucleotide
(b) Nucleus
(c) Nucleoid
(d) Nucleosome
B) TRUE/ FALSE:
6) Translation is the last step of Central Dogma.
7) Histone octamer is negatively charged.
8) Genetic code is triplet.
C) FILL UPS:
9) Central dogma was proposed by......
10) Adenine and Guanine are.......
ANSWER KEY: PART- A |
A) MULTIPLE CHOICE QUESTIONS:
1) (a) N- glycosidic bond
2) (c) DNA synthesis
3) (b) Translation
4) (c) Transcription
5) (b) Nucleosome
B)TRUE/ FALSE:
6) True
7) False
8) True
C) FILL UPS:
9) Francis Crick
10) Purines
PART: B SHORT ANSWER TYPE QUESTIONS:
11) How nucleoside is different from nucleotide?
12) Define Genetic Code.
13) Define Nucleosome.
PART:C LONG ANSWER TYPE QUESTIONS:
14) Differentiate between —
(a) Replication and Transcription
(b) Transcription and Translation.